There has been a lot interest on simulation the devolpment of infectious deseases since the COVID-19 pandemic of 2019/20. Many different models and approaches exist to computationally simulate the spread of infectious deseases.
Among these the compartmental models and especially the quite simple SIR model have been around since the late 1920s and successfully applied to epidemics resp. pandemenics around the world.
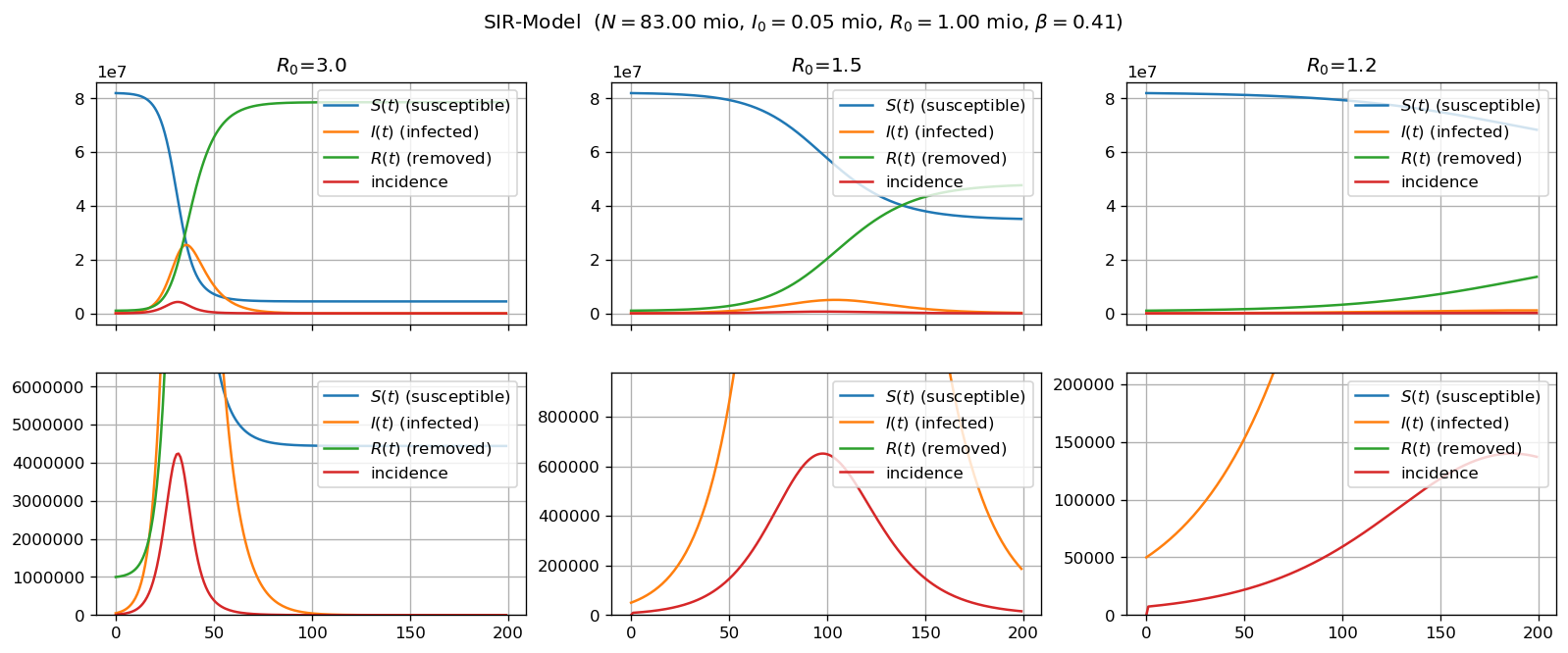
The SIR-model is a classical model in the sense that it is described as a system of three ordinary differential equations. As such it is very descriptive and can quite easily be motivated and understood. I wrote a Jupyter notebook which (a) tries explain the approach of the SIR-model and (b) implements a simple python simulation of the model.
You can find the python notebook here and the sources in github repository here.